植物分子生理学研究室
Lab for Plant Molecular Physiology, Fac Appl Biol Sci, Gifu University
research area
2.Analysis of Transcriptional Network at the Heart of Environmental Adaptation of Higher Plants
Sessile plants have evolved excellent ability to adapt to a wide range of environmental conditions for their survival. The heart of the ability is the transcriptional response to environmental change that is implemented by the transcriptional network of higher plants. Currently, most of the network is hidden in a veil of mystery. We are trying to reveal this network through several approaches, including bioinformatics, functional genomics, and biochemistry.
keywords:
environmental response, large-scale expression
analysis, NGS, CAGE, RNAseq, promoter prediction, AlphaScreen, transcriptional network, signal crosstalk, TF relay, cis-trans pairing, systems biology.
3.Transcriptional Response to Light Stress
Light is necessary for plants to grow as the energy source of photosynthesis, but excess light damages photosynthetic apparatus, chloroplasts, and tissues, inhibits plant growth and threatens plant’s life. In addition, light stress can be observed at other stress conditions, such as cold, UV-B, drought, and nutrition-deficient stresses. We are trying to understand these stresses in a view of light stress. Methods of population genetics (GWAS) are also utilized.

keywords:
light stress, cold stress, UV-B stress, photoinhibition,
chlorophyll fluorescence, PAM, photosynthetic
electron transfer, state transition, ROS, redox
regulation, photoprotection, ELIP, phenylpropanoid
biosynthesis,
4.Promoter Structure and Evolution
Plant Promoters are not static in the nucleotide sequence. Thanking to the great power of NGS, evolution of plant promoters are studied in order to understand genetic adaptation of plants to environments. Methods of population genetics (GWAS) are also utilized.
keywords:
regulatory SNP, transposon, GWAS, local adaptation, ecogenomics.
5.Photosynthetic Seaslug
Some seaslugs in sacoglossa are able to photosynthesize by a phenomenon called “kleptoplasty”.
keywords:
secondary endosymbiosis, kleptoplasty, seaslug, green algae.
keywoords
OUR RESEARCH
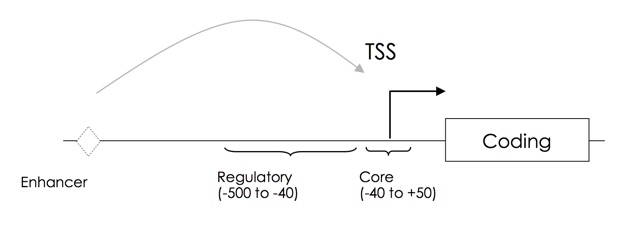
1.Genome-Wide Promoter Analysis
In our lab, computational and experimental analyses are combined to elucidate plant promoter structure. Synthetic promoters are also utilized to validate predicted cis-regulatory elements and to understand the transcriptional network in higher plants. Some of the results can be accessed at our plant promoter database (ppdb, http://ppdb.agr.gifu-u.ac.jp).

keywords:
TSS, NGS, CapTrapper, CAGE, LDSS, in silico analysis,
promoter structure, core promoter, transcriptional
regulatory element.
topics:
Heterogeneity of Plant Core Promoter









































Highly Cited Researchers of Gifu University




